Cython Tutorial
I have always been loving Python. When people mention Python, they usually think of the following two advantages:
- Easy to write
- Easy to call C/C++ libraries
In fact, the first bullet point comes with super slow execution speed. And the second bullet point requires C/C++ libraries to export APIs according to Python specification, which might not be available for most of the libraries.
During the internship at Tianrang Inc., I had several experiences with Cython. I think although this tool is buggy sometimes and has terrible user experience sometimes, it can boost Python significantly and also help create wraps for C/C++ libraries. In this article, I’d introduce Cython to readers. Notice that, Cython is different from CPython. Cython helps us to conveniently:
- Use Python syntax to mix code in C/C++ and Python while boosting Python performance
- Call C/C++ libraries
Example: Matrix Multiplication
Suppose we are writing some computation-heavy code, for example, Matrix Multiplication. Remember that this case is meant to show you how easy it is to boost Python performance using Cython, not discussing how to optimize matrix multiplication which is a non-trivial task.
Let’s assume matrices store in numpy.ndarray. Cython can treat ndarray as continuous memory space thus accessing should be very fast. We can write the matrix multiplication like this:
# dot_python.py
import numpy as np
def naive_dot(a, b):
if a.shape[1] != b.shape[0]:
raise ValueError('shape not matched')
n, p, m = a.shape[0], a.shape[1], b.shape[1]
c = np.zeros((n, m), dtype=np.float32)
for i in xrange(n):
for j in xrange(m):
s = 0
for k in xrange(p):
s += a[i, k] * b[k, j]
c[i, j] = s
return c
It looks quite simple. You can even imagine how it will be like if you write it in C/C++, just almost the same except for the syntax. Of course, we would expect it to run slower than the C/C++ version. So how do we use Cython to optimize it? As simple as this:
# dot_cython.pyx
import numpy as np
cimport numpy as np
cimport cython
@cython.boundscheck(False)
@cython.wraparound(False)
cdef np.ndarray[np.float32_t, ndim=2] _naive_dot(np.ndarray[np.float32_t, ndim=2] a, np.ndarray[np.float32_t, ndim=2] b):
cdef np.ndarray[np.float32_t, ndim=2] c
cdef int n, p, m
cdef np.float32_t s
if a.shape[1] != b.shape[0]:
raise ValueError('shape not matched')
n, p, m = a.shape[0], a.shape[1], b.shape[1]
c = np.zeros((n, m), dtype=np.float32)
for i in xrange(n):
for j in xrange(m):
s = 0
for k in xrange(p):
s += a[i, k] * b[k, j]
c[i, j] = s
return c
def naive_dot(a, b):
return _naive_dot(a, b)
We can see that the Cython code looks almost the same as the Python code. Let me explain the difference:
- Cython program has extension
.pyx cimportis used in Cython to import.pxdfiles. You can think.pxdfiles as header files in Cython. The two headers we imported here are built-in.@cython.boundscheck(False)and@cython.wraparound(False)two decorators are used to turn off boundary checks.-
Use
cdefto define Cython functions. We can specify the types of arguments and the return value. For example, here is how we can write theminfunction for integers:cdef int my_min(int x, int y): return x if x <= y else yBack to our matrix multiplication program,
np.ndarray[np.float32_t, ndim=2]is a type name likeint, but it’s longer and contains more information. It says the variable is anp.ndarrayarray whose elements have the type ofnp.float32_tand whose dimension is two. - Inside a function body, we can define variables like
cdef typename varname. - Cython functions won’t be exported to Python world. So we have
def naive_dot(a, b)to wrap up_naive_dotCython function.
Also, Cython programs need to compile. Here is the big picture of how it works:
- Cython compiler compiles Cython code to C/C++ code that calls Python APIs and exports APIs according to Python specification.
- The generated C/C++ code compiles to a dynamic library.
- Python interpreter loads the dynamic library.
To finish the first two steps, we need to have:
# setup.py
from distutils.core import setup, Extension
from Cython.Build import cythonize
import numpy
setup(ext_modules = cythonize(Extension(
'dot_cython',
sources=['dot_cython.pyx'],
language='c',
include_dirs=[numpy.get_include()],
library_dirs=[],
libraries=[],
extra_compile_args=[],
extra_link_args=[]
)))
It seems too complicated for our simple example, but the good news is that it cannot be more complicated, even for very complicated programs. Let me explain those parameters:
'dot_cython'is the name of the target dynamic library.sourcesis a list of source files including.pyx,.cand.cppfiles.languagedefaults toc. If you need to use C++, simply change it toc++.include_dirsis passed to-Iarguments ofgcclibrary_dirsis passed to-Larguments ofgcclibrariesis passed to-larguments ofgccextra_compile_argsis passed togcc. For example, you might want to-std=c++11or-DSOME_MACRO.extra_link_argsis passed togcc.- If you are not familiar with those
gccarguments, it’s completely okay. You will know them when you need them.
After writing this setup.py, we can use the following command to compile the Cython program to a dynamic library:
python setup.py build_ext --inplace
After this, we will find two extra files in the current directory: dot_cython.c and dot_cython.so. The former is the generated C program and the latter is the dynamic library.
Let’s see the performance:
$ ipython 15:07:43
Python 2.7.12 (default, Oct 11 2016, 05:20:59)
Type "copyright", "credits" or "license" for more information.
IPython 4.0.1 -- An enhanced Interactive Python.
? -> Introduction and overview of IPython's features.
%quickref -> Quick reference.
help -> Python's own help system.
object? -> Details about 'object', use 'object??' for extra details.
In [1]: import numpy as np
In [2]: import dot_python
In [3]: import dot_cython
In [4]: a = np.random.randn(100, 200).astype(np.float32)
In [5]: b = np.random.randn(200, 50).astype(np.float32)
In [6]: %timeit -n 100 -r 3 dot_python.naive_dot(a, b)
100 loops, best of 3: 560 ms per loop
In [7]: %timeit -n 100 -r 3 dot_cython.naive_dot(a, b)
100 loops, best of 3: 982 µs per loop
In [8]: %timeit -n 100 -r 3 np.dot(a, b)
100 loops, best of 3: 49.2 µs per loop
You can see that the Cython code is 570x faster and we didn’t change much of the code. Of course, it’s still slower than numpy which is highly optimized. Again, we don’t care about matrix multiplication optimizations. We just want Python to run as fast as C/C++. How about writing the same matrix multiplication algorithm in C/C++? Here is the example:
// dot.cpp
#include <ctime>
#include <cstdlib>
#include <chrono>
#include <iostream>
class Matrix {
float *data;
public:
size_t n, m;
Matrix(size_t r, size_t c): data(new float[r*c]), n(r), m(c) {}
~Matrix() { delete[] data; }
float& operator() (size_t x, size_t y) { return data[x*m+y]; }
float operator() (size_t x, size_t y) const { return data[x*m+y]; }
};
float dot(const Matrix &a, const Matrix& b) {
Matrix c(a.n, b.m);
for (size_t i = 0; i < a.n; ++i)
for (size_t j = 0; j < b.m; ++j) {
float s = 0;
for (size_t k = 0; k < a.m; ++k)
s += a(i, k) * b(k, j);
c(i, j) = s;
}
return c(0, 0); // to comfort -O2 optimization
}
void fill_rand(Matrix &a) {
for (size_t i = 0; i < a.n; ++i)
for (size_t j = 0; j < a.m; ++j)
a(i, j) = rand() / static_cast<float>(RAND_MAX) * 2 - 1;
}
int main() {
srand((unsigned)time(NULL));
const int n = 100, p = 200, m = 50, T = 100;
Matrix a(n, p), b(p, m);
fill_rand(a);
fill_rand(b);
auto st = std::chrono::system_clock::now();
float s = 0;
for (int i = 0; i < T; ++i) {
s += dot(a, b);
}
auto ed = std::chrono::system_clock::now();
std::chrono::duration<double> diff = ed-st;
std::cerr << s << std::endl;
std::cout << T << " loops. average " << diff.count() * 1e6 / T << "us" << std::endl;
}
$ g++ -O2 -std=c++11 -o dot dot.cpp
$ ./dot 2>/dev/null
100 loops. average 1112.11us
We can see that the Cython performance is the same as the hand-written C++ code.
The Key to Performance Boost
What if we get rid of all type annotations in the Cython code, i.e. types of function arguments and the return value, and also those cdef in the function body? Here is the result:
$ python setup.py build_ext --inplace
$ ipython
In [1]: import numpy as np
In [2]: import dot_python
In [3]: import dot_cython
In [4]: a = np.random.randn(100, 200).astype(np.float32)
In [5]: b = np.random.randn(200, 50).astype(np.float32)
In [6]: %timeit -n 100 -r 3 dot_cython.naive_dot(a, b)
100 loops, best of 3: 416 ms per loop
In [7]: %timeit -n 100 -r 3 dot_python.naive_dot(a, b)
100 loops, best of 3: 537 ms per loop
We can see that the Cython code without type annotation is as slow as the Python code. Therefore, type annotation is the key to performance boost.
That’s a significant disadvantage of dynamically typed languages. Lots of information that could have been determined at compile time will have to postpone until runtime. Lots of optimization that could have been done at compile time will have to postpone until runtime as well. What’s worse, CPython does not have a Just-in-Time compiler. Thus lots of time is wasting on type checking, not to mention lots of compilation optimizations.
Analyse Cython Programs
We know that type annotation is the key to the performance boost. But we also see that without type annotation it will still be a valid Cython program, but not efficient. How do we avoid forgetting to add type annotation? Cython provides a great tool that points out parts that could potentially further optimize.
cython -a dot_cython.pyx
If the Cython program uses C++, remember to add --cplus argument. cython -a generates a .html file with the same filename. Let’s see what it tells us about the non-annotated version:
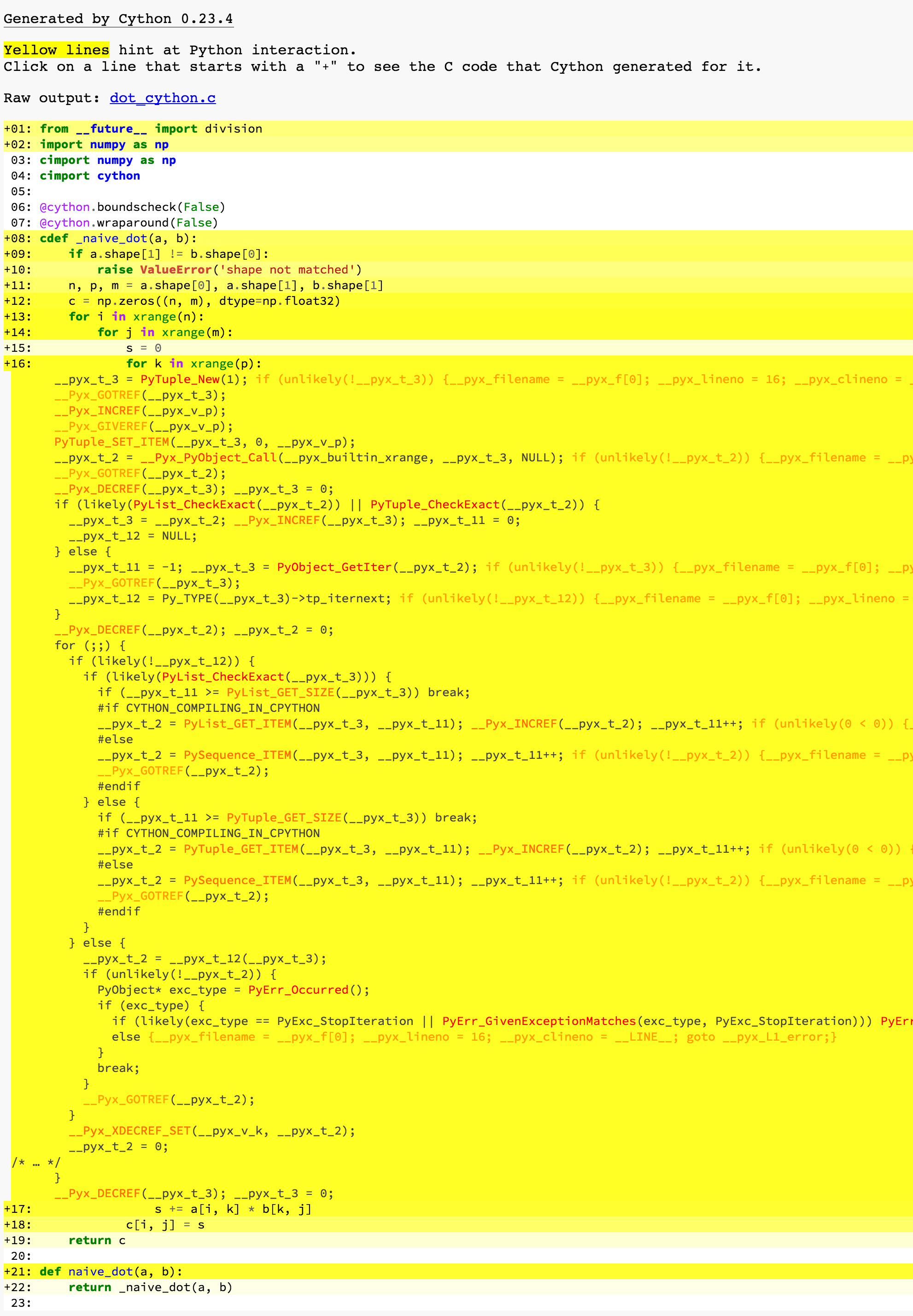
Yellow parts are where the generate C/C++ code calls Python APIs. You can simply regard it as what makes the program slow. Clicking a line will show the corresponding generated C/C++ code. We can find that almost all lines are marked yellow, no wonder it is so slow.
See Line 16, for k in xrange(p). It is supposed to be a simple for-loop, but the generated code is so complicated. By looking at the name of those Python APIs, the program does lots of extra works, such as creating and destroying PyObject, increment and decrement the reference counting of PyObjects, type checking, boundary checking, and so on. Unfortunately, if you don’t know what types those variables are in advance, you will have to do all that extra work.
How about we add type annotations back?
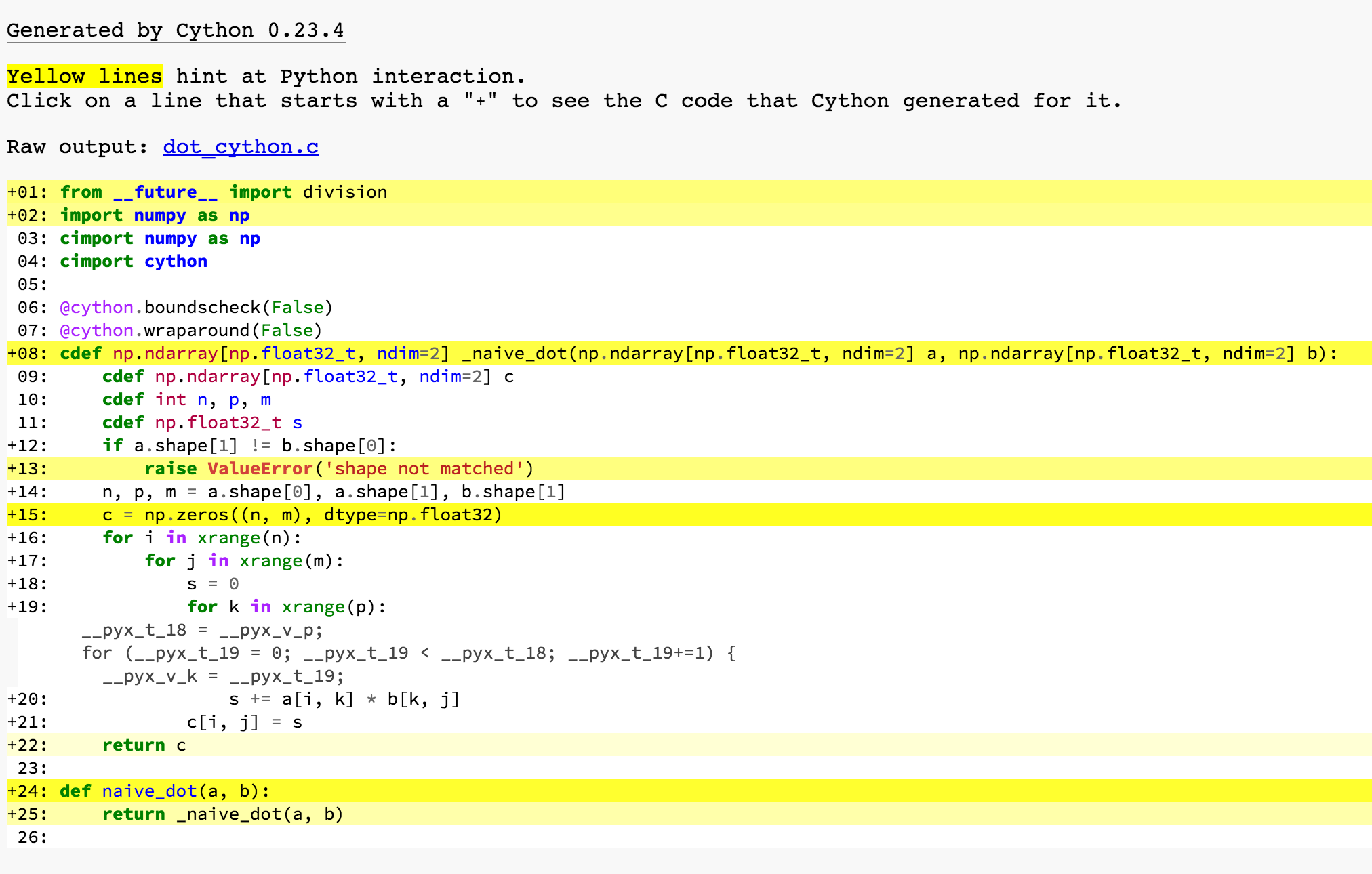
It looks much better. Let’s look at for k in xrange(p) again. The generated code is a simple for-loop, as we expected. The rest yellow parts are invoking the function and returning, raising exceptions, and calls np.zeros which is a Python function. Those are the places where you have to call Python APIs.
According to Amdahl’s Law and also by instinct, we know that as long as the most frequently visited code is fast enough, the whole program will be fast enough. Therefore, we can enjoy the simplicity of Python and rewrite the core using Cython or C/C++. That gives you both performance and development productivity.
Here are some references to the above section:
- Cython for NumPy users
- Faster code via static typing
- Dynamic type languages versus static type languages
- Language Basics
Glue C/C++ Together with Python
Python is a great glue language, but the libraries need to call Python APIs and follow Python specifications. With the help of Cython, we can easily wrap a C/C++ library for Python to call.
I wanted to write this section in detail like the performance section, but then I realized it’s all about Cython syntax. All you need is Cython documentation. Let me briefly list some features:
- Function signature is almost the same as C/C++
_Boolin C andboolin C++ are replaced withbint
struct/enum/unionare supportedconstand references are supported- Namespaces are supported
- C++ classes are supported
- Most of operator overloadings are supported. Some of them require name changes.
- Nested classes are supported
- Templates are supported
- Exceptions are supported
- Constructor and destructor are supported
- Static members are supported
libc/libcpp/ STL are supported- Use
cimport to import.pxdfiles in.pyx` files - Mind the convertion between Python string and all kinds of C/C++ strings
To sum up, it should be okay to call C/C++ functions in Cython call. And it’s also okay to write C/C++ programs using Python syntax. To know more, you might want to read: